View description
Putative protein of unknown function; null mutant is viable and displays elevated frequency of mitochondrial genome loss; null mutation confers sensitivity to tunicamycin and DTT
Localization:
Intensity:
Fold change:
Significance:
-
C’ GFP library in SD
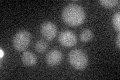
below threshold15.39 -
N' NOP1pr-GFP in SD

below threshold12.1047 -
N' TEF2pr-mCherry in SD

below threshold4.7894 -
N' NATIVEpr-GFP in SD
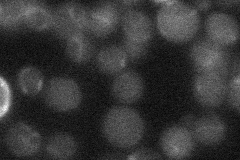
below threshold18.8044 -
N' TEF2pr-VC and Cyto-VN in SD

#N/A0 -
C’ GFP library in SD+DTT

cytosol16.81.09No -
C’ GFP library in SD+H2O2
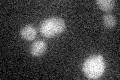
cytosol16.771.08No -
C’ GFP library in Starvation Media

cytosol15.330.99No -
C’ GFP library on the background of Pup2-DaMP

below threshold -
C’ GFP library on the background of CCT mutant

below threshold14.83990.963313No
