Sphingolipid complexity and fine-tuning
Although the idea that cell membrane lipid bilayers do little more than give shape and form to cells and limit diffusion between cells and their environment is totally passé, the structural, compositional, and functional complexity of lipid bilayers often catches cell and molecular biologists by surprise. Models of lipid bilayer structure have developed considerably since the heyday of the fluid mosaic model, principally by the discovery of the restricted diffusion of membrane proteins and lipids within the plane of the bilayer. We recently suggested that further refinement of current models is necessary and proposed that describing lipid bilayers as “finely-tuned molecular assemblies” best portrays their complexity and function.
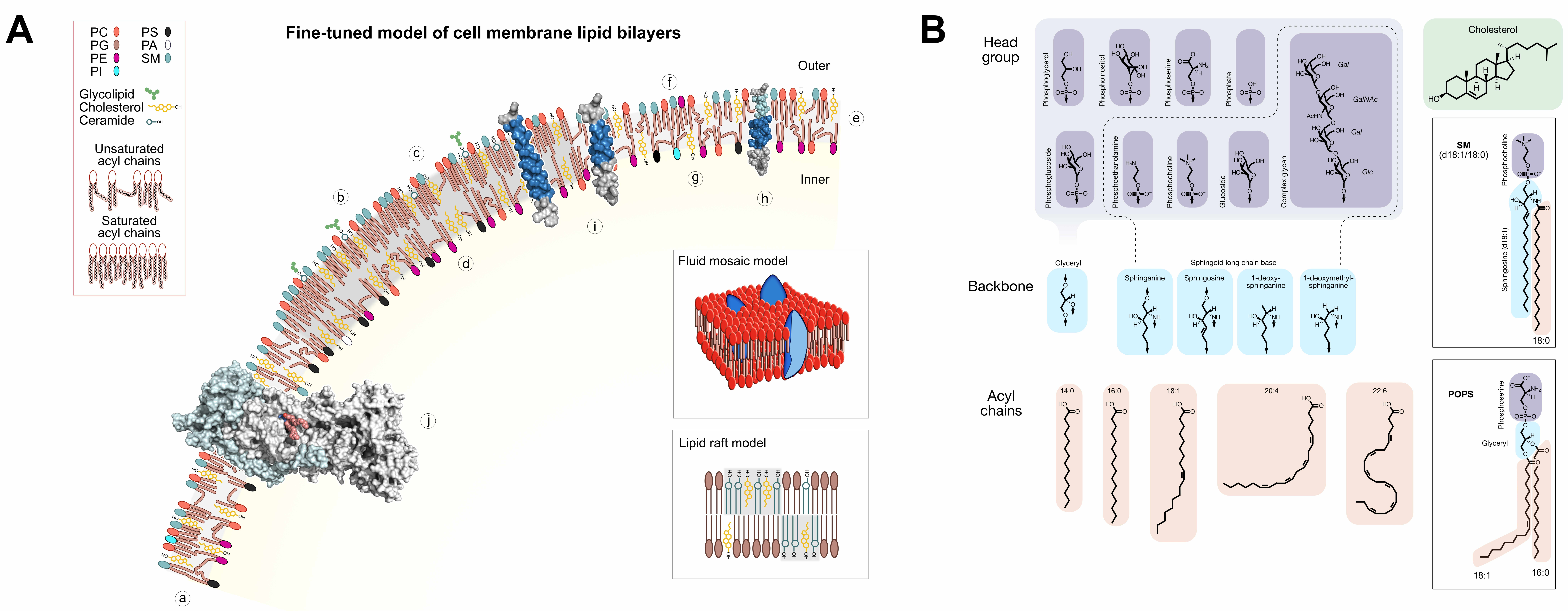
Figure 1. Panel A) Evolution of models of the lipid bilayer culminating in the proposal that lipid bilayers are best described as finely tuned molecular assemblies. While the fluid mosaic model (middle right) of the 1970s portrayed a homogenous sea of lipids (red) with mobile protein islands (blue), the lipid raft model (lower right) introduced the concept of lateral heterogeneity. Rafts are enriched in saturated sphingolipids (brown), ceramide (blue), and cholesterol (yellow), have tighter lipid packing, and increased hydrophobic height. The current model that we now propose depicts the multiple fine-tuned features of lipid bilayers; a list of these features (a–j) is indicated in the scheme. The model, showing 156 individual lipid molecules (a), was generated using recent analysis of the composition and topology of lipids in an erythrocyte plasma membrane bilayer. The key to each lipid species is given in the inset in the upper left. The asymmetric packing of the bilayer leaflets is shown (c): more tightly packed lipids with unsaturated acyl chains in the outer leaflet, and (d) fewer well-packed lipids containing unsaturated acyl chains found at higher levels on the inner leaflet. Bilayer width is indicated in gray, darker shading corresponding to domains of restricted lateral mobility (b). One lipid transporter protein (P4-ATPase bound to palmitoyloleoylphosphatidylserine; PDB ID: 6k7m) is indicated (j) along with transmembrane domains of generic proteins (i and h) indicating asymmetry in their surface area that matches the difference in lipid packing (g) between leaflets. PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin.
Panel B) Eukaryotic membranes consist of three major lipid classes, namely glycerolipids, sphingolipids, and sterols. The structural complexity of the first two classes is indicated by the range of headgroups (purple), backbones (blue), and acyl chains (orange) that combine to yield the cellular lipidome. The head groups normally found in sphingolipid structures are indicated by a dashed line. Two examples of typical lipid structures are shown on the right (boxes), where SM (N-stearoyl-D-erythro-sphingosylphosphorylcholine) consists of a d18:1 sphingoid long-chain base backbone to which a C18:0 fatty acid is N-acylated, and PS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine), which consists of a glycerol backbone to which two different fatty acids are esterified, one of which is saturated (16:0) and one unsaturated (18:1).
Figure panels reproduced from: Dingjan, T., & Futerman, A.H. (2021). The fine‐tuning of cell membrane lipid bilayers accentuates their compositional complexity. BioEssays, 43, 2100021–2100021. https://doi.org/10.1002/bies.202100021
The complexity of cell membranes is exemplified by analysis of one of their key components, namely sphingolipids. Sphingolipids can be differentiated from other membrane lipids by the distinctive chemistry of the sphingoid long chain base (LCB), which is generated by the condensation of an amino acid (normally but not always serine) and a fatty acyl CoA (normally palmitoyl CoA) by the pyridoxal phosphate-dependent enzyme, serine palmitoyl transferase (SPT). The first five carbon atoms of the sphingoid LCB, which we defined as the ‘sphingoid motif’, are largely responsible for the unique chemical and biophysical properties of sphingolipids since they can undergo a relatively large number (compared to other lipid species) of molecular interactions with other membrane lipids, via hydrogen-bonding, charge-pairing, hydrophobic and van der Waals interactions. These interactions are responsible, for instance, for the association of sphingolipids with cholesterol in the membrane lipid bilayer. The remarkable selectivity of the fine-tuned interactions of sphingolipids within the lipid membrane bilayer, and the subtle means by which these interactions are modified and regulated in eukaryotic cells, raise a number of challenging questions about the generation of these proteins, and of their interactions with the sphingoid motif in evolutionary history.
The lab is currently studying, by a combination of in silico and experimental work, how these finely tuned interactions are regulated, and attempting to understand their significance in membrane and cell function.
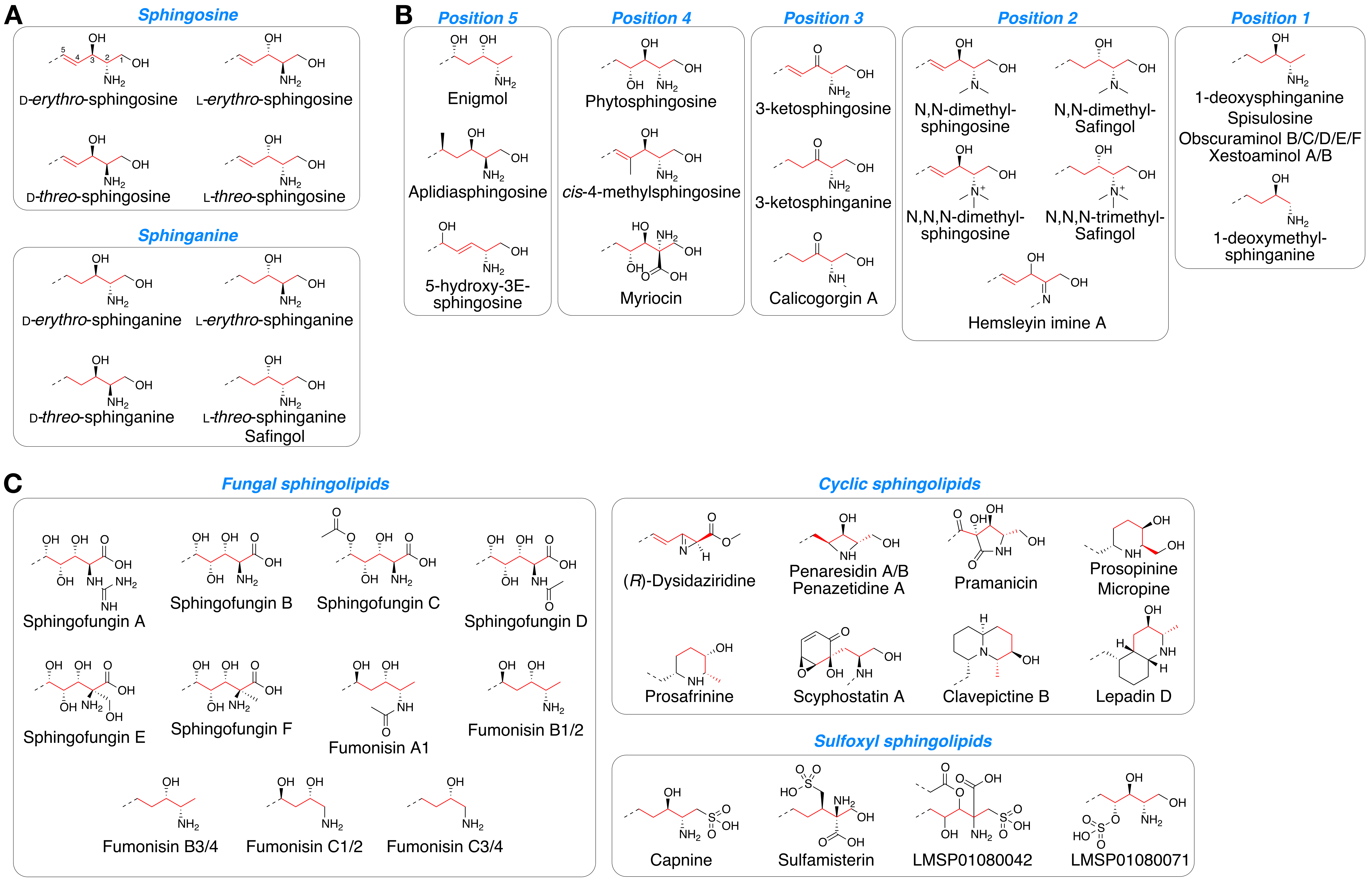
Figure 2. The sphingoid motif and selected substitutions. The structures, taken from the LIPID MAPS structure database, portray a range of substitutions in the sphingoid motif (red), but exclude substitutions that generate more complex SLs, such as N-acylation (to form ceramides), O-phosphorylation (to form SM), and O-glycosylation (to form glyco-SLs). Panel A) Variations to the stereochemistry of the sphingoid motif; canonical sphingolipids are defined by a d-erythro stereoconfiguration. Panel B) substitutions at each of the 5 positions of the sphingoid motif gives rise to a range of natural sphingolipid-derived lipid species. Panel C) Common structural modifications of sphingolipids include hydroxylation (often found in fungal sphingolipids), cyclisation, and sulfonylation.
Figure reproduced from: Dingjan, T., & Futerman, A.H. (2021). The role of the ‘sphingoid motif’ in shaping the molecular interactions of sphingolipids in biomembranes. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1863, 183701. https://doi.org/10.1016/j.bbamem.2021.183701
