RNA cis-regulatory elements (Riboregulators), such as riboswitches, play a significant role in gene expression regulation in bacteria. These elements are transcribed upstream to a gene and control its expression by sensing a specific ligand in the cell. The ligand could be a certain metabolite, tRNA, the ribosome itself or a protein, depending on the system type. Upon binding the ligand, the riboregulator changes its structure leading to alteration of the expression of the downstream genes. Many riboregulators work at the transcription level: generating a conditional transcription terminator or anti-terminator upon ligand binding.
PASIFIC aims to identify these alternative conformations and assess the probability of them being a functional riboregulator.
For more information please see: Millman et al. 2016
Source code for PASIFIC is available at
Please note this tool works best in Chrome browser
Strat by entering up to 10 sequences of suspected riboregulators in FASTA format:
PASIFIC works best when you are confident about the termination site, so suspected sequences must end at the poly-U site of the terminator. If you do not know the exact 3' end of the transcript check the option for 3' prediction:

When his option is checked, PASIFIC will scan the whole sequence for a terminator.
Optimization:
There are several search parameters that can be changed, and optimize the search for suspected system type: attenuator, riboswitch or t-box. Choose one of the system types or change the parameters manually to your preferences. Click the "Default" link to return to default parameters

Please note that the classification data set was folded using default parameters, hence the probability score with altered parameters is less reliable.
FASTA format:
The results page shows the color coded alternative conformations of the input sequence, both in dot-bracket notation and as downloadable figures (svg format). Color codes:
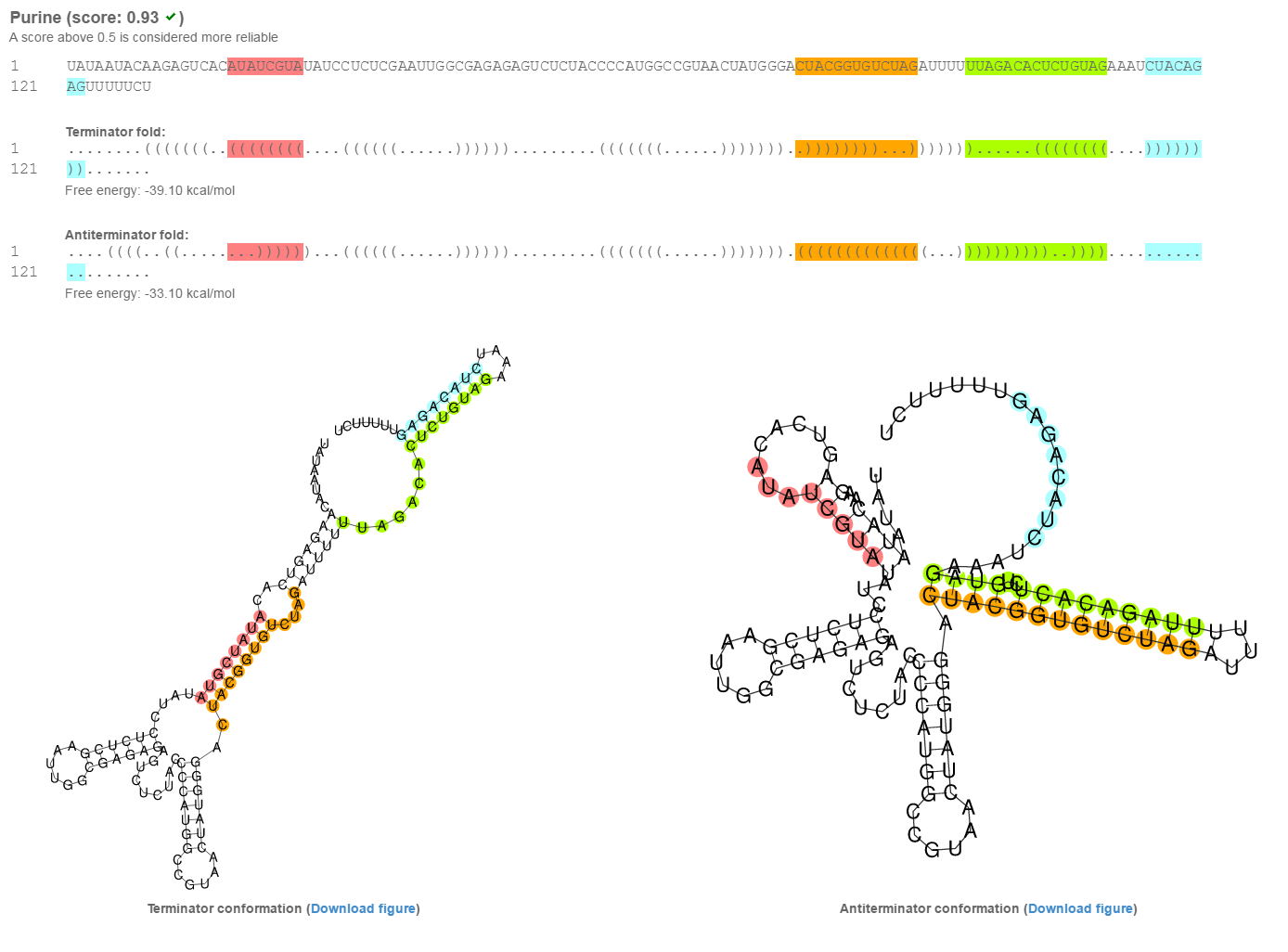
Next to the sequence title, in parentheses, is the probability score. A threshold of 0.5 yields sensitivity of 82.5% and specificity of 80.6%. For more details please turn to the paper.