Reading RNA modifications
Since the discovery of RNA modifications in the 1950’s, the field had relied on approaches interrogating modifications one by one. We sought to leverage the advances in sequencing in order to systematically profile RNA modifications at a transcriptome wide scale. The main challenge we had to overcome is that Illumina sequencing based approaches rely on sequencing of DNA. Once RNA is reverse transcribed into DNA, the information regarding the modification is typically lost. To overcome this challenge, we developed a suite of approaches - often relying on unique chemistries - allowing for information regarding the presence of a modification to be relayed also following reverse transcription. In some cases, we employed chemistries that cause an RNA modification to be improperly reverse transcribed, leading to accumulation of mutations in the DNA. In other cases, the modifications cause reverse transcriptase to abort at the modified site, leading to an accumulation of DNA fragments that all terminate at the same sites. In other cases, we employed affinity reagents allowing to ‘fish’ modified fragments.
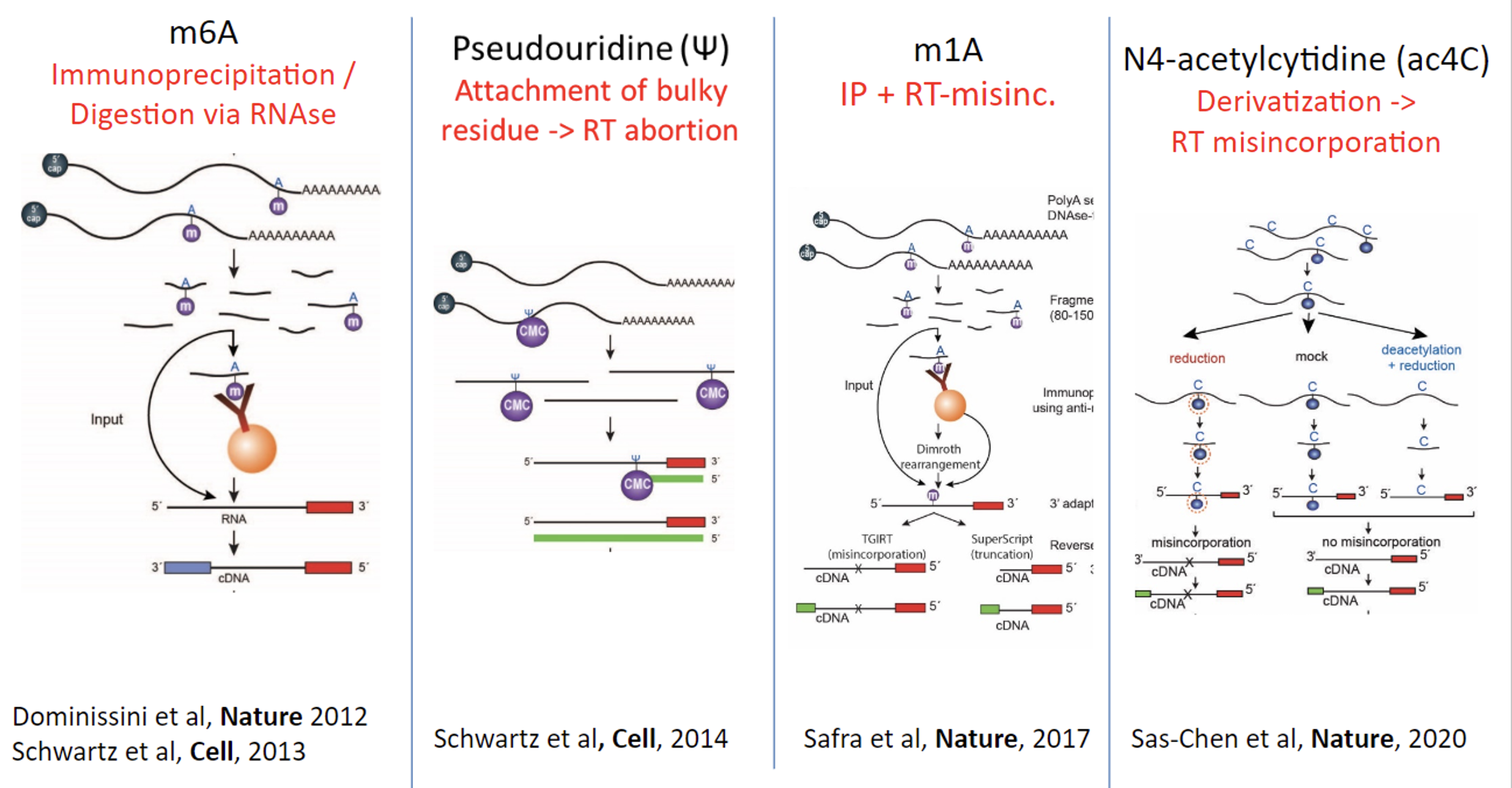
These approaches allowed us to chart the RNA epitranscriptome. We uncovered the first maps of diverse modifications, across different model species. These maps served as a critical stepping stone for uncovering the enzymatic machineries governing their formation, understanding the sequence and structure based code giving rise to the specific deposition of modifications at individual sites, for exploring contexts in which different modifications are dynamically regulated, for characterizing the functional impact of modifications on mRNA stability and translation, and for studying how RNA modifications evolve.
Publications
- Dynamic RNA acetylation revealed by cross-evolutionary mapping at base resolution
Sas-Chen A., Thomas J. M., Matzov D. et al., (2020) Nature. 583, p. 638–643 - The m(1)A landscape on cytosolic and mitochondrial mRNA at single-base resolution
Safra M., Sas-Chen A., Nir R. et al. (2017) Nature. 551, 7679, p. 251-255 - Transcriptome-wide Mapping Reveals Widespread Dynamic-Regulated Pseudouridylation of ncRNA and mRNA
Schwartz S., Bernstein D. A., Mumbach M. R. et al. (2014) Cell. 159, 1, p. 148-162 - High-Resolution Mapping Reveals a Conserved, Widespread, Dynamic mRNA Methylation Program in Yeast Meiosis
Schwartz S., Agarwala S. D., Mumbach M. R. et al. (2013) Cell. 155, 6, p. 1409-1421 - Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S. et al. (2012) Nature. 485, 201–206
