Understanding RNA modifications
We are highly interested in understanding the roles played by different RNA modifications, and how alterations in RNA modifications give rise to disease. A key modification we have focused on is adenosine methylation (m6A), the most widespread modification on mRNA, present at tens of thousands of sites within human mRNAs. Recently, we were able to establish that m6A serves as the key factor governing mRNA decay. It has long been unclear why different genes are subject to decay at varying levels. Our findings revealed that m6A - installed in the nucleus, in a manner dependent on splicing - serves as a covalent mark that dictates cytoplasmic decay.
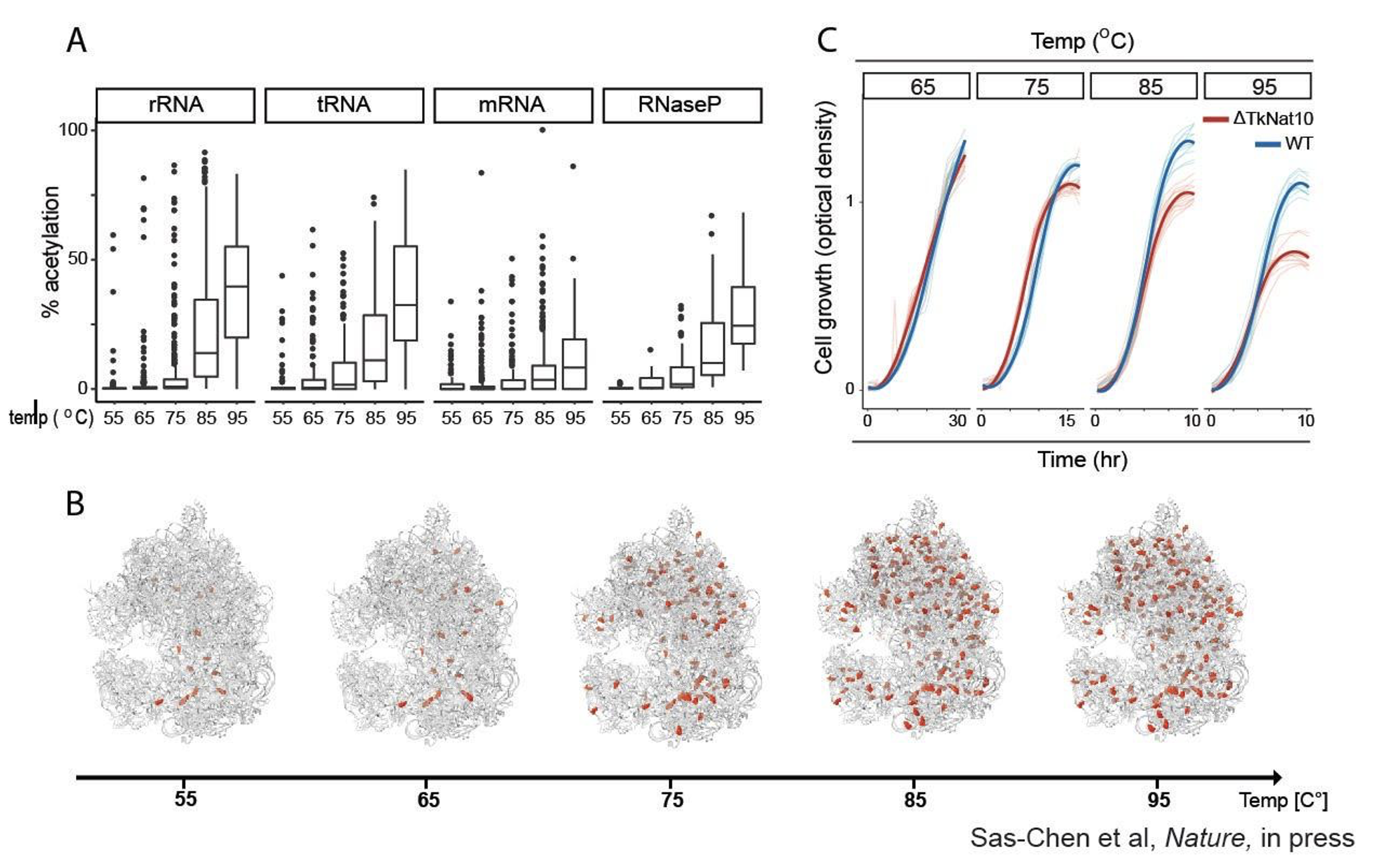
A different modification we have been working on is cytidine acetylation (ac4C). We discovered that in the course of adapting to higher temperatures, ribosomal RNA (rRNA) and tRNA of the hyperthermophile T. kodakarensis, an archaeal extremophile that thrives at ultra-high degrees of up to 100℃, reshaped themselves and acquired an unprecedented number of acetylated residues at ~200 target sites across the ribosome. We further found that RNA acetylation was required for growth at higher, but not lower, temperatures. What is the role played by ac4C? We found that ac4C - when introduced in an RNA duplex - results in more stable duplexes. A key challenge hyperthermophiles need to address is how to allow highly structured RNAs (such as rRNA or tRNA) to remain stable at high temperatures, in which such structures are typically denatured. Our findings highlight one solution to this problem, in the form of modifying their building blocks from cytidines to acetylated cytidines (Nature, 2020). This study opened up an exciting ongoing ERC-funded direction in our lab, of uncovering the roles and dynamics of other RNA modifications of the ribosomal machinery, unraveling the biological contexts in which such regulation occurs and understanding how their dynamic modulation controls ribosome structure and function.
Publications
- Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability
Uzonyi A., Dierks D., Nir R. et al. (2023) Molecular Cell. 83, 2, p. 237-251.e7 - Dynamic RNA acetylation revealed by cross-evolutionary mapping at base resolution
Sas-Chen A., Thomas J. M., Matzov D. et al., (2020) Nature. 583, p. 638–643
