The immune system of bacteria
Bacteria, the most abundant organisms on the planet, are outnumbered by a factor of 10 to 1 by phages that infect them. Faced with the rapid evolution and turnover of phage particles, bacteria have evolved various mechanisms to evade phage infection and killing, leading to an evolutionary arms race (Ofir & Sorek, Cell 2018). We study this arms race in order to understand how the extensive co-evolution of both phage and host shapes the huge diversity of the microbial world (Bernheim et al, Nature Reviews Microbiology 2020).
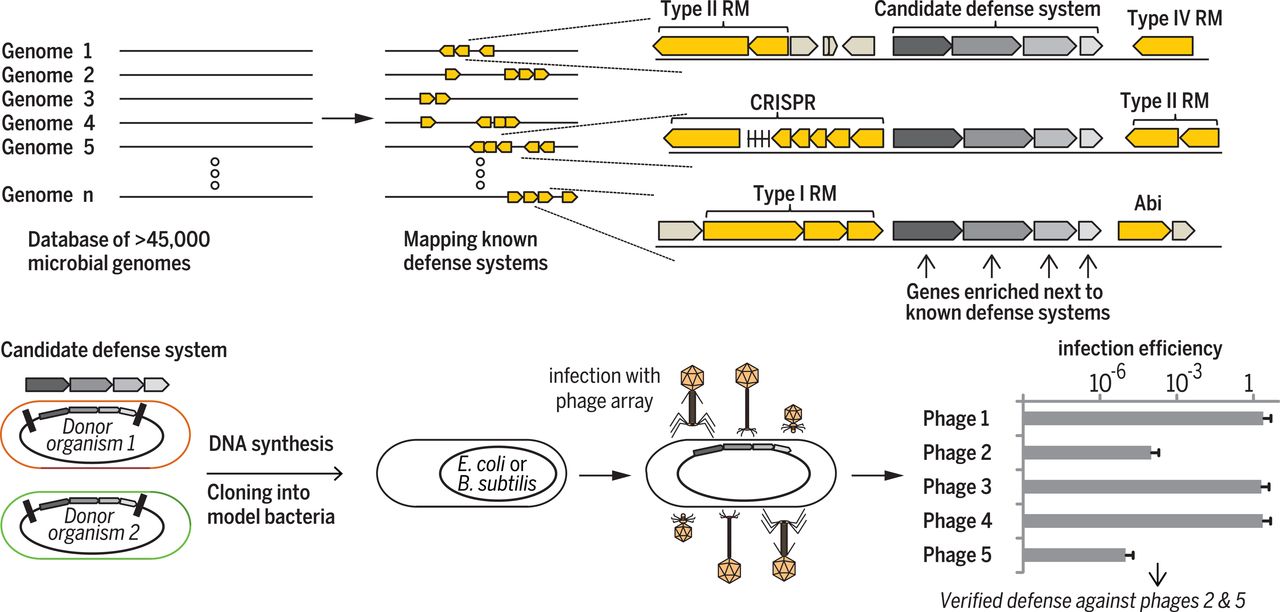
After a few years studying CRISPR-Cas , the adaptive immune system of bacteria (Levy et al, Nature 2015), we realized that bacteria encode a rich and diverse set of defense systems that was, until our studies, obscure. Our studies rely on the observation that bacterial defense systems cluster together in specific genomic loci called “defense islands”, which we genomically mine to discover new defense systems (Doron et al, Science 2018).
Within bacterial defense islands, our studies revealed over 50 new defense systems, encoding a plethora of defense mechanisms demonstrating surprising complexity in the bacterial immune system. For example, we found bacteria encoding defense systems that use new kinds of small molecules for intracellular signaling (Cohen Nature 2019; Tal Cell 2021; Ofir Nature 2021; Leavitt Nature 2022; Rousset Science 2025); found bacteria that use reverse-transcribed non-coding RNAs to mediate defense against phage (Millman Cell 2020); and characterized giant immune complexes that are 4 times larger than the ribosome (Duncan-Lowey, Cell 2023). We also showed that some bacterial immune systems can produce a variety of small molecule inhibitors of viral replication (Bernheim, Nature 2021), described how bacteria can deplete essential cellular molecules as a measure of antiviral immunity (Tal, Nature Microbiology 2022; Garb, Nature Microbiology 2022; Rousset, Cell 2023), and demonstrated how some defense systems protect against phage by interfering with virion assembly (Hör, Nature 2024).
A surprising and unexpected aspect of our findings was the realization that important components of the human innate immune system evolved from defense systems that protect bacteria from phage infection (Wein, Nature Reviews Immunology 2022). We showed that the human cGAS-STING antiviral pathway (Cohen Nature 2019; Morehouse, Nature 2020), Toll-interleukin receptor (TIR) immune signaling (Doron, Science 2018; Ofir Nature 2021; Leavitt Nature 2022), Viperin antiviral proteins (Bernheim, Nature 2021), the Pyroptosis inflammatory pathway (Johnson Science 2022; Wein Nature 2025), and caspase-mediated immune cell death (Rousset Science 2025) all originated in bacteria where they serve to defend against phage. In addition to explaining how our immune system evolved, we demonstrated how understanding mechanisms in the bacterial immune systems can solve new immune mechanisms in animals and plants (e.g., Ofir Nature 2021; Leavitt Nature 2022; Rousset, Cell 2023).

