Magnetic Resonance Spectroscopic Fingerprinting
 NMR can detect signals from small molecules in the brain. The relaxation times of these molecules (basically, the time it takes them to return to thermal equilibrium when perturbed from it) are excellent markers of a large range of pathologies in the brain.
NMR can detect signals from small molecules in the brain. The relaxation times of these molecules (basically, the time it takes them to return to thermal equilibrium when perturbed from it) are excellent markers of a large range of pathologies in the brain.
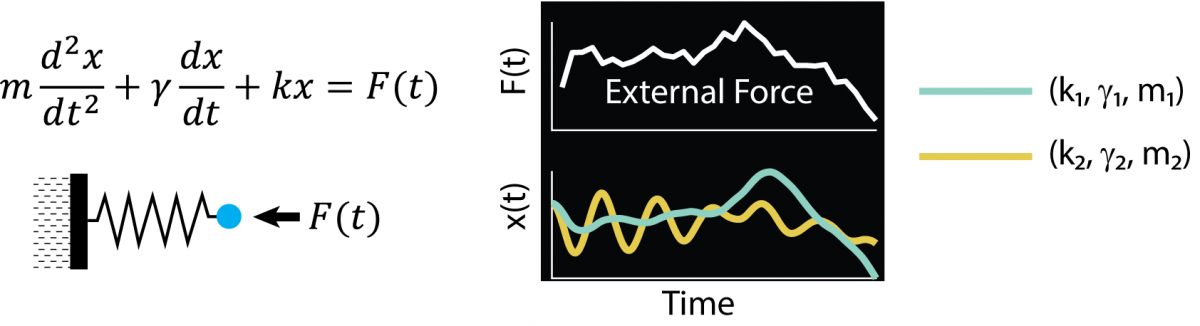
We have recently developed a new and fast method for measuring the relaxation times of metabolites, by applying a “semi-random” series of radiofrequency pulses to the brain. Much like the shaking of an opaque box to try and understand what’s inside, the pulses cause spins with different parameters to evolve differently in quantum-mechanical Liouville space. We can track the evolution of the spins, which creates a temporal “fingerprint”. The fingerprint “encodes” the relaxation times of the metabolites. To use a simple analogy, consider a harmonic oscillator: by applying a specially designed driving force, the oscillator will move back and forth in a quasi-random manner. The trajectory x(t) will depend on the system’s spring constant (k), damping constant (g) and mass (m). By observing the temporal evolution, you can try and deduce the oscillator’s constants.
Our research focuses on understanding how to best model the temporal evolution of the spins, on designing novel irradiation patterns which encode the maximal amount of information into our fingerprints, and in using such fingerprinting approaches to undersample and accelerate our measurements.
Adaptive Spectroscopy
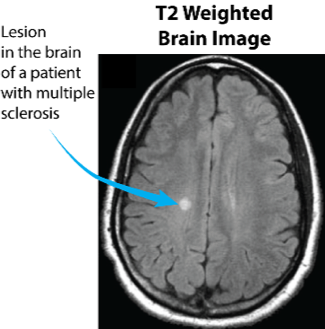 Imagine trying to measure the elasticity of a ball, by throwing it multiple times on the ground with different amounts of force and watching it bounce. It might seem obvious to you that you should vary your force in response to the ball’s dynamics: if it barely bounces, you should increase the amount of force, while if it bounces too much, you should diminish it. This means that you adapt yourself to the experiment’s results in real time. It might seem obvious to you, but in MRI and NMR, such approaches for designing experiments have not yet been explored, and we’re exploring their potential impact. Our focus is on measuring relaxation times of spins (T1, T2), which are powerful biomarkers for many diseases. For example, multiple sclerosis is a neurodegenerative disease in which lesions form inside the brain. The image to the right maps the T2 relaxation time of protons in water, and shows it changes in such a manner that allows for the detection of lesions.
Imagine trying to measure the elasticity of a ball, by throwing it multiple times on the ground with different amounts of force and watching it bounce. It might seem obvious to you that you should vary your force in response to the ball’s dynamics: if it barely bounces, you should increase the amount of force, while if it bounces too much, you should diminish it. This means that you adapt yourself to the experiment’s results in real time. It might seem obvious to you, but in MRI and NMR, such approaches for designing experiments have not yet been explored, and we’re exploring their potential impact. Our focus is on measuring relaxation times of spins (T1, T2), which are powerful biomarkers for many diseases. For example, multiple sclerosis is a neurodegenerative disease in which lesions form inside the brain. The image to the right maps the T2 relaxation time of protons in water, and shows it changes in such a manner that allows for the detection of lesions.
Changing the experiment adaptively raises many fascinating questions: How do you design your experiment in the first place? How to set the experimental parameters? How to change them adaptively, as information is collected in real time? The answers to these questions borrow deeply from information theory, statistics and machine learning, and sit at the interface between physics, engineering, computer science and mathematics.
Motor Learning
 We study the neurochemical changes that occur in the brain in response to motor learning. Learning occurs when one neuron excites repeatedly the following neuron. This is called Hebbian learning, and it leads to long term potentiation, in which additional synapses are formed between the two neurons. When motor learning takes place, it has long been hypothesized that a reduction in neuronal inhibition from pre-motor neurons is necessary to allow for increased excitability. We have shown that a reduction of GABA, the brain’s major inhibitory neurotransmitter, occurs in the motor cortex shortly after learning (~10-30 minutes), and that this reduction is correlated with the learning rate.
We study the neurochemical changes that occur in the brain in response to motor learning. Learning occurs when one neuron excites repeatedly the following neuron. This is called Hebbian learning, and it leads to long term potentiation, in which additional synapses are formed between the two neurons. When motor learning takes place, it has long been hypothesized that a reduction in neuronal inhibition from pre-motor neurons is necessary to allow for increased excitability. We have shown that a reduction of GABA, the brain’s major inhibitory neurotransmitter, occurs in the motor cortex shortly after learning (~10-30 minutes), and that this reduction is correlated with the learning rate.
 Multiple questions await to be answered: how do these gains change over time? When and why does the inhibitory system normalize? How is this related to changes in brain connectivity? What behavioral aspects of motor learning trigger the change? And what other exciting things can we learn by translating our work to the newly acquired 7 Tesla MRI, which offers us unprecedented resolution and sensitivity.
Multiple questions await to be answered: how do these gains change over time? When and why does the inhibitory system normalize? How is this related to changes in brain connectivity? What behavioral aspects of motor learning trigger the change? And what other exciting things can we learn by translating our work to the newly acquired 7 Tesla MRI, which offers us unprecedented resolution and sensitivity.