We assemble on-chip compartments using a photo-activable biocompatible monolayer for immobilizing DNA polymers brushes coding for genes expressed in a cell-free reaction. Our studies exposed entropy-driven collective conformations in a brush as a function of its density and ionic strength, and revealed osmotic, salted, neutral and mushroom phases in DNA predicted by Phil Pincus 25 years ago for general polyelectrolyte brushes. We further observed that DNA brushes create a compartmentalized environment with brush length, density, and promoter orientation creating an effective boundary and macromolecular composition differences. In recent work, we directly measured the accumulation of RNA polymerases, ribosomes, nascent RNAs and proteins, in dense DNA brushes, with transcription and translation coupled in the brush. The colocalization of machinery and nascent RNA and proteins to the brush creates favorable conditions for coupling machine synthesis and assembly.
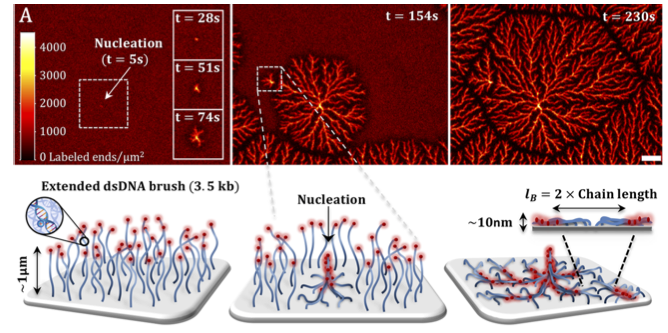
We investigated the response of DNA brushes to multi-valent cations and discovered exotic phases of condensed DNA. An extended osmotic brush undergoes a first order transition into dendritic structures of collapsed condensed DNA, which nucleate locally and grow into macroscopic 2D domains separated by grain boundaries. Bridging of cations compensates for the electrostatic repulsion between DNA chains, minimizing the free energy by these structures. Recently, we realized that electron beam lithography applied to our photo-activable monolayer can be used to immobilize DNA polymers on patterns much thinner (100nm) than the length of the DNA (1 µm), thereby forming a line of single DNA polymers – a 1D brush. These brushes then condense into 1D structures of bundles of ~30 co-aligned DNA molecules, roughly 20nm in diameter, and hundreds of microns long. DNA condensation in 1D propagating in a network graph patterns could solve computational problems, such as finding the minimal path in a 2D representation of a maze.
Buxboim, M. Bar-Dagan, V. Frydman, D. Zbaida, M. Morpurgo and R. Bar-Ziv. A single-step photolithographic interface for cell-free gene expression and active biochips. Small, (3):500-10, (2007).
S.S. Daube, D. Bracha, A. Buxboim, R. Bar-Ziv. Compartmentalization by directional gene expression. Proc. Natl. Acad. Sci. USA 107(7):2836-2841 (2010).
D. Bracha, E. Karzbrun, G. Shemer, P. Pincus, R. Bar-Ziv. Entropy-driven collective interactions in DNA brushes on a biochip. Proc. Natl. Acad. Sci. USA, 110 (12), 4534-4538 (2013).
D. Bracha and R.Bar-Ziv. Dendritic and Nanowire Assemblies of Condensed DNA Polymer Brushes, J. Am. Chem. Soc., 136(13) 4954-4953 (2014).
D. Bracha, E. Karzbrun, S.S. Daube, R. Bar-Ziv. Emergent properties of dense DNA phases toward artificial biosystems on a surface, Acc. Chem. Res., 47 (6), 1912–1921 (2014).
G. Pardatscher, D. Bracha, S.S. Daube, O. Vonshak, F.C. Simmel, R. Bar-Ziv. DNA condensation in one dimension, Nature Nanotechnology. 11, 1076-1081, (2016).
M. Levy, R. Falkovich, O.Vonshak, D.Bracha, A.M. Tayar, Y.Shimizu, S.S.Daube, R. Bar-Ziv. Boundary-Free Ribosome Compartmentalization by Gene Expression on a Surface, ACS Synth. Biol., 10, 3, 609–619 (2021).