-
Prof. Uri Alon


 Systems biology and physics of behavior
Systems biology and physics of behaviorUnderstanding the protein circuits that perform computations within the cell is a central problem in biology. In physics, cells offer the challenge of understanding the collective behavior of interacting molecular machines designed to operate with remarkable precision under strong biological constraints. Our lab studies biological circuits, and applies the principles we discover also to understand dynamics on a different scale, that of human interactions.
- Network motifs: basic interaction patterns that recur throughout biological networks, much more often than in random networks
- Development of dynamic proteomics, a system for monitoring the position and amounts of endogenous proteins in individual living human cells
- We study evolution when multiple objectives are at play. When a biological system needs to perform more than one task, it faces a fundamental tradeoff, which leads to surprising simplicity in the range of phenotypes
- Theatre lab: we use concepts from improvisation theatre, and tools from physics and computer science to study basic principles of human interactions
-
Prof. Ayelet Erez


 deciphering the dynamics of cellular metabolism at different disease states.
deciphering the dynamics of cellular metabolism at different disease states.Our lab deciphers the dynamics of cellular metabolism in different disease states. In particular, we are interested in understanding the contribution of the urea cycle components to the metabolic changes accompanying disease pathogenesis. Urea cycle metabolites serve as metabolic junctions that directly contribute to distinct metabolic fluxes at different cellular states. Consequently, changes in the function of urea cycle proteins have important effects on cell growth, survival, and proliferation. Identifying specific metabolic alterations during disease can improve diagnosis, progress monitoring, and therapeutic interventions.
-
Prof. Tamar Geiger


 Clinical Cancer Proteomics
Clinical Cancer ProteomicsThe interplay between cancer genetic aberrations and external microenvironmental cues is the basis of tumorigenic growth and metastasis. Despite extensive research, the internal heterogeneity of tumours presents enormous challenges and limits our ability to achieve complete therapeutic responses. Although tumour heterogeneity has been thoroughly investigated at the genomic and transcriptomic levels, limited studies have investigated heterogeneity at the proteomic level. We study tumor heterogeneity using cutting-edge mass spectrometry-based proteomics of cancer clinical samples, and investigate the relation between the cancer cells and the immune system within the tumor microenvironment. We develop novel single cell proteomic approaches, and combine them with tumor analyses and bioinformatics to characterize the cellular interactions within the tumor microenvironment, and how these interactions associate with response to immunotherapy. We further investigate the association between the proteomic level and the genomic and transcriptomic levels aiming to elucidate gene-expression regulatory mechanisms and the functional proteomic output of cancer somatic mutations.
-
Prof. Shalev Itzkovitz
Department Head


 Systems Biology of Mammalian Tissues
Systems Biology of Mammalian TissuesWe combine mathematical modelling and sensitive single-cell measurements to study the design principles of mammalian tissues. Specific topics include:
- Homeostatic mechanisms of intestinal stem cells
- Design principles of the mammalian liver
- Intercellular interactions in pancreatic islets
- Single molecule approaches for studying gene expression in intact tissues
-
Prof. Valery Krizhanovsky


 Cellular senescence in tissue damage cancer and aging
Cellular senescence in tissue damage cancer and agingSenescent cells accumulate in premalignant lesions, sites of tissue damage and in normal tissues during aging. We study how senescent cells affect cancer, aging and age-related diseases and also placenta during embryonic development.
- Cellular senescence in aging and age-related diseases
- Cellular senescence in premalignant lesions and cancer
- Interaction of senescent cells with the immune system
- Cellular senescence during embryonic development
-
Prof. Sima Lev


 Cell Signaling in Cancer Development and Metastasis
Cell Signaling in Cancer Development and Metastasis- Signaling networks in triple negative breast cancer (TNBC) subtypes
- Combination therapy, drug resistance, and cancer stem cells
- RTKs (AXL, cMet, EGFR) and non-RTKs (PYK2, FAK) signaling in TNBC
- Ferroptosis and TNBC therapy
- Epithelial-mesenchymal transition (EMT) & TNBC metastasis
-
Prof. Gil Levkowitz


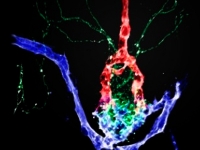 Linking genes, development and function of the zebrafish hypothalamus
Linking genes, development and function of the zebrafish hypothalamusOur lab utilizes zebrafish as a vertebrate model organism to tackle basic questions concerning the development and function of the hypothalamus. The hypothalamus is an evolutionarily ancient and conserved brain region that allows all vertebrates to adapt to emotional and physiological challenges. Hypothalamic neurons regulate fundamental body functions including sleep, blood pressure, body temperature, hunger and thirst, stress and social behavior.
Our main goals are to identify the molecular mechanisms underlying:
- Morphogenesis and cell biology of the hypothalamo-neurohypophyseal system (NHS), which is an important neuroendocrine conduit between brain and blood through which neurohormones are released from hypothalamic neurons into the general circulation without disrupting the blood-brain barrier.
- The effects of early developmental processes on physiological functions of the mature hypothalamus, focusing on stress response and social behavior.
-
Prof. Elior Peles


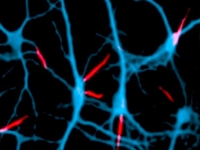 Development of myelinated nerves
Development of myelinated nervesResearch in the lab focuses on the biology of Schwann cells and oligodendrocytes, the myelinating glial cells of the peripheral and central nervous system, respectively. We are using a variety of molecular, biochemical and genetic approaches to:
- Identify the axonal signals that control myelination
- Understand how glial cells recognize and wrap axon with myelin
- Dissect the cytoskeletal machinery that drives myelination
- Reveal how myelinating glia shape the membrane of the axons they wrap
-
Prof. Yardena Samuels


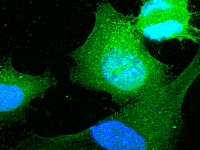 Melanoma Functional Genomics
Melanoma Functional GenomicsOur group aims to discover recurrent tumor-specific mutations in melanoma using whole exome and whole genome sequencing approaches. Once mutations are identified, our group focuses on characterizing the biochemical, functional, and clinical aspects of the most highly mutated genes. To facilitate this search, we have established a unique bank of metastatic melanoma tumors which is used for:
- In depth functional analysis of melanoma mutations using novel preclinical models such as somatic cell knockout and somatic cell knockin isogenic cells
- Identification of melanoma protein/protein interactions using overexpression and endogenous epitope tagging,” (EET) approaches. This will uncover which signal transduction pathways mutated proteins impinge upon
- Delineate clinically ideal melanoma protein targets using single and combination drug screens
- In addition to the potential insights into basic tumor biology, new opportunities for clinical intervention are likely to be generated through this research
-
Prof. Oren Schuldiner


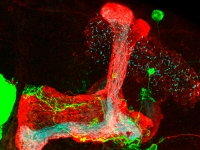 Mechanisms of neuronal remodeling
Mechanisms of neuronal remodelingOur lab studies the molecular mechanisms that regulate, control and execute developmental neuronal remodelling in Drosophila. Remodelling is essential for sculpting the mature nervous systems of both vertebrates and invertebrates during development and when it is perturbed, we think it is a major cause of psychoneurological conditions such as schizophrenia and autism.
We use Drosophila as a model system to study pruning and regrowth during remodelling and our main approach is developmental genetics. The stereotypy of remodeling in Drosophila as well as the wide range of genetic tools available, make it a unique system to dissect the mechanisms of neuronal remodeling in vivo. -
Prof. Eran Segal


 Developing Computational Models for Personalized Medicine
Developing Computational Models for Personalized MedicineHeads a multi-disciplinary team of computational biologists and experimental scientists working in the area of Computational and Systems biology. Focuses on Nutrition, Genetics, Microbiome, and Gene Regulation and their effect on health and disease. His lab aims to develop personalized nutrition and personalized medicine using machine learning, computational biology, probabilistic modeling, and analysis of heterogeneous high-throughput genomic and clinical data.
-
Prof. Liran Shlush



We study the aging of the human hematopoietic system and how age related clonal hematopoiesis associates with inflammation and aging. We study leukemia in its early staged and looking for new strategies to target preleukemia. We try to understand how the aging of the human bone marrow environment influences the blood system.
-
Prof. Yonatan Stelzer


 Epigenetics in development and disease
Epigenetics in development and diseaseEpigenetic modifications provide cells and organisms with remarkable plasticity. Yet, disentangling the Gordian knot of epigenetic cause and effect still remains a formidable task. Building on the recent developments in single cell genomics, pluripotent stem cells, and genome editing and moving forward, by developing systems for dissection of epigenetic function in-vivo, represents a deep and fundamental challenge our group is determined to engage. Specific interest includes:
- How are cell-specific epigenetic programs established in the early embryo? How do they modulate cell state and function?
- What are the effects of epimutations on development and disease?
- Can environmental factors influence the germline epigenome - potentially affecting subsequent generations?
-
Prof. Ravid Straussman


 The study of resistance to anti-cancer therapy
The study of resistance to anti-cancer therapyWe use cutting-edge, high-throughput technologies to study the different mechanisms that render cancer cells resistant to anti-cancer therapy. Our goal is to advance anti-cancer precision medicine for the creation of better treatment options for cancer patients. Topics of interest include:
- Tumor microenvironment-mediated chemoresistance
- The human microbiome and chemoresistance
- Cell autonomous rewiring as a mechanism for chemoresistance
- The study of drug tolerant persisters
- Study of the timing and mechanisms that drive genetic chemoresistance
- Common models: Melanoma, Breast cancer, lung cancer, pancreatic cancer
-
Prof. Amos Tanay

 Making sense of biological and medical data
Making sense of biological and medical dataOur group is a hub of cutting edge biological and medical data. We combine computational and experimental work to understand how single cells regulate their genes, enhancers and chromosomal conformations. We develop models to test how cells work together to form (artificial) tissues in new embryonic models, and we combine single cell work and analysis of massive electronic health records to explore how cells within tissues lose stability during age-related disease. Most importantly - we are passionate about understanding how biology works and believe massive data analysis and mechanistic/causal understanding should never be in conflict - their synthesis represents not just one of the greatest, but also one of the most fun challenges in science.
-
Prof. Itay Tirosh


 Tumor Systems Biology
Tumor Systems BiologyThe Tirosh lab is studying human tumors as a complex ecosystem in which diverse cancer and non-cancer cells interact and collectively determine tumor biology and response to therapies. We leverage single cell technologies, computational approaches and clinical collaborations to identify important tumor subpopulations such as cancer stem cells, drug resistant cells, invasive cells and immune cells that respond to immunotherapies. We then study the function and regulation of these subpopulations in model systems, with the ultimate goal of developing better cancer treatments.
-
Prof. Eldad Tzahor


 Cardiomyocyte cell-cycle control and dedifferentiation
Cardiomyocyte cell-cycle control and dedifferentiationThe Tzahor lab focuses on regenerative medicine, with particular relevance to adult heart diseases. Heart disease is the number one cause of death, surpassing the number of deaths from all cancers combined. Its burden on society and healthcare systems is immense. Our lab's inspiring insights into mechanisms of cardiac regeneration have led to fundamental shifts in biomedical knowledge and have positioned us among the top labs in the field of cardiac regeneration. We revealed that, contrary to common belief, latent regeneration machinery can be awakened even in the adult heart (Tzahor 2017, Science).
Current projects demonstrating this notion are briefly summarized below:
- ErbB2-Yap crosstalk in heart regeneration
- Agrin therapy for heart disease
- Regenerative senescence in the heart
- Copaxone as a novel drug for heart disease
- Fibroblasts-Immune cells crosstalk
- Ion channels and mechano-sensory regulation of cardiomyocytes
- Cardio-Oncology
- Myoblast fusion
-
Dr. Leeat Yankielowicz-Keren


 Systems tissue pathology
Systems tissue pathologyThe Keren lab uses MIBI-TOF (Multiplexed Ion Beam Imaging by Time of Flight), a novel imaging platform in which antibodies conjugated to metals are used to simultaneously visualize dozens of proteins by Secondary Ion Mass Spectrometry (SIMS). The result is a high-dimensional image, depicting sub-cellular protein expression and localization in situ.
Emeriti
-
Prof. Avri Ben-Zeev

 Adhesion-mediated signaling during cancer progression
Adhesion-mediated signaling during cancer progressionWe study the changes in the mechanisms that regulate the coordination between cell-cell adhesion and signaling during cancer invasion and metastasis. Special emphasis is devoted to the following topics:
- The dual role of beta-catenin in WNT signaling (as co-transcription activator) and as a key mediator of cell-cell adhesion linking adhesion receptors to the cytoskeleton.
- Identification and role of WNT/beta catenin target genes in regulating cell motility, epithelial to mesenchymal transition (EMT), and cancer cell invasion and metastasis.
- Investigation of genes induced by adhesion-mediated (WNT and others) signaling that are also expressed at increased levels in normal stem cells and during cancer progression. We are studying the role of such genes in both tissue homeostasis and cancer progression.
-
Prof. Eli Canaani

Investigates the MLL gene and its protein products, in order to understand their biochemical activities and the mechanism by which they trigger leukemia. Leukemias associated with rearrangement of the MLL gene account for the majority of acute lymphocytic and myelocytic leukemias in infants, and in therapy-related leukemias.
-
Prof. Zvi Kam


We develop and apply methodologies of Light Microscope Image Acquisition and of Quantitative Analysis to cell biology. Studies with collaborators, to which these methodologies are applied include:
- Dynamic behavior of osteoclasts sealing zone (Prof. Benny Geiger & Dr. Sarit Batsir).
- Individual and collective cell migration in 2D and 3D environments. (such as computer analysis of wound-healing essay, work of Yair Elisha and Prof. Benny Geiger)
- Platelets spreading and activation.
- Lamelapodia dynamics.
- Robust algorithm for tracking cells (Figure: the NetFlow graph optimization for tracking cells in time-lapse movies, proposed by Prof. Adi Shamir)
-
Prof. Moshe Oren


 Molecular and cellular biology of cancer
Molecular and cellular biology of cancerOur aim is to understand the molecular and cellular processes that control normal cell (and tissue) behavior, and how they become perturbed in cancer. Specifically, we focus on:
- The p53 tumor suppressor pathway
- The Hippo tumor suppressor pathway
- Chromatin modifications in cancer (particularly histone H2B ubiquitination)
- microRNAs in cancer
- Crosstalk between cancer cells and their microenvironment
-
Prof. Varda Rotter

 The role of p53 in maintaining genomic plasticity
The role of p53 in maintaining genomic plasticity- Mutant p53 gain of function
- p53 Loss of Heterozygosity (LOH)
- Role of p53in the life of stem cells
- Role of p53 in the life of cancer stem cells
- p53 in inflammation metabolism
- p53 regulating endocrine loops
- p53 based therapy
-
Prof. Yehiel Zick

 Cross talk between insulin resistance, animal lectins and bone remodeling
Cross talk between insulin resistance, animal lectins and bone remodelingWe combine mouse models, cell biology and analysis of signaling pathways to focus on:
- Role of animal lectins (particularly galectin-8) as regulators of bone remodeling and insulin action
- Novel elements (Ndfip1; Otub2, TM7SF3) that modulate survival of pancreatic beta cells
- IRS (insulin receptor substrate) proteins, insulin resistance, and beta cell function























































































































































































































































