Systems Protobiology - Studies of the Origin of Life
In the realm of prebiotic evolution, we have proposed a Lipid World model for life’s emergence, involving primordial assemblies of lipid-like amphiphilic molecules, which can faithfully pass their compositional information to progeny. We have devised an artificial chemistry formalism, the Graded Autocatalysis Replication Domain (GARD) model to test our hypothesis. The model describes, by computer simulations, the chemical dynamics of simple prebiotic molecules in environments of large molecular repertoires. GARD depicts the biased accretion kinetics of molecular assemblies that are kept far from equilibrium by occasional fission events. This biased accretion leads to the spontaneous emergence of homeostatic growth, underlying self-reproduction of these assemblies.
Our simulations demonstrate that by simply implementing nature-like mutually-catalytic interactions between accreting molecules, forming metabolism-like networks, random assemblies of such molecules progressively attain the capacity to self-reproduce. We use the term "composome" to refer to an assembly that maintains compositional homeostasis over growth-and-split generations, and the term "compotype" to an area in compositional space that is populated by highly-similar composomes. The spontaneous proliferation of lipid assemblies, and the transfer of their compositional genome information to progeny, constitute a primitive self-reproduction mechanism, much simpler than the ones involving templating biopolymers (i.e. RNA-First). This suggests that self-copying of individual molecules, such as RNA, commonly taken as a unique prerequisite for life’s emergence, may be replaced by mutually-catalytic networks with effective overall system’s evolvability.
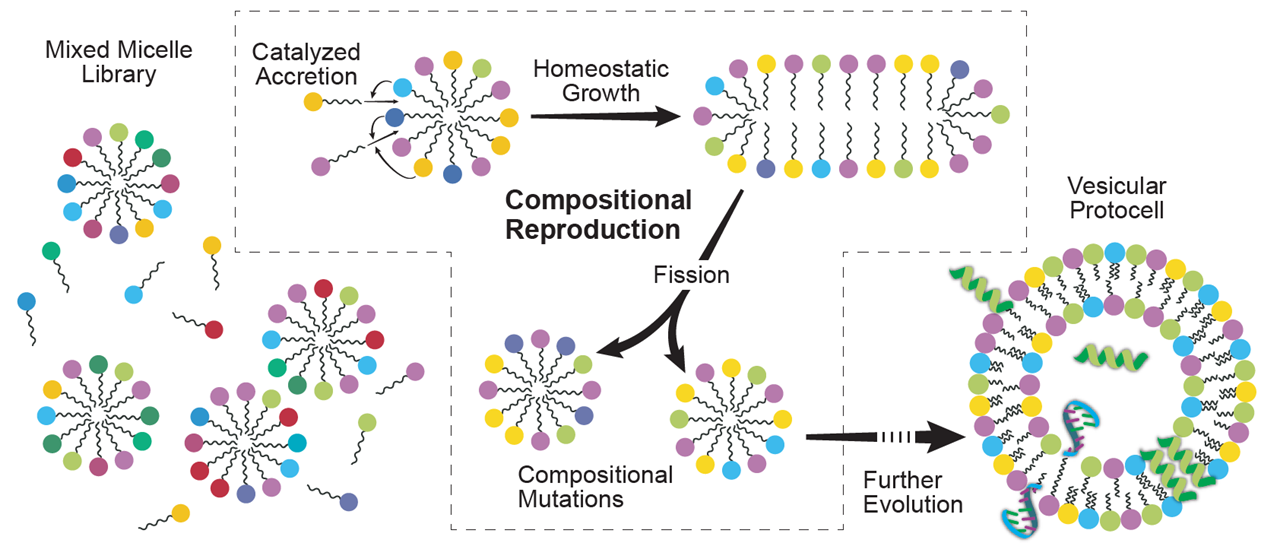
Origin of Life in self-reproducing catalytic micelles
In our recent projects, we have diversified our approaches to accumulate new evidence to the foundational principles of the Lipid-First scenario. For example, we examined reports of the aqueous organic molecular repertoire of Saturn's moon Enceladus, and compared it to analyses of advanced Mass-Spectrometry data of primordial meteorites and other organic sources, showing the prevalence and dominance of amphiphiles in prebiotic settings. We expanded our simulation capacities to explore lipid accretion kinetics in Molecular Dynamics, an advanced computational tool reliably simulating the dynamics of molecular systems in atomistic and picosecond resolutions. Lastly, we continue to explore and expand the GARD model, highlighting the significant Origin of Life implications of its emergent properties.
Selected Publication
- Kahana, A., & Lancet, D. (2021). Dynamic lipid aptamers: non-polymeric chemical path to early life. Chemical Society Reviews (In Press).
- Kahana, A., & Lancet, D. (2021). Self-reproducing catalytic micelles as early nanoscopic protocells. Nature Reviews Chemistry (In Press).
- Lancet, D., Segrè, D., & Kahana, A. (2019). Twenty years of “lipid world”: a fertile partnership with david deamer. Life, 9(4), 77.
- Kahana, A., Schmitt-Kopplin, P., & Lancet, D. (2019). Enceladus: First observed primordial soup could arbitrate origin-of-life debate. Astrobiology, 19(10), 1263-1278.
- Kahana, A., & Lancet, D. (2019). Protobiotic systems chemistry analyzed by molecular dynamics. Life, 9(2), 38.
- Lancet, D., Zidovetzki, R., & Markovitch, O. (2018). Systems protobiology: origin of life in lipid catalytic networks. Journal of The Royal Society Interface, 15(144), 20180159.
- Gross, R., Fouxon, I., Lancet, D., & Markovitch, O. (2014). Quasispecies in population of compositional assemblies. BMC evolutionary biology, 14(1), 1-11.
- Markovitch, O., & Lancet, D. (2014). Multispecies population dynamics of prebiotic compositional assemblies. Journal of theoretical biology, 357, 26-34.
- Markovitch, O., & Lancet, D. (2012). Excess mutual catalysis is required for effective evolvability. Artificial life, 18(3), 243-266.
- Kafri, R., Markovitch, O., & Lancet, D. (2010). Spontaneous chiral symmetry breaking in early molecular networks. Biology direct, 5(1), 1-13.
- Inger, A., Solomon, A., Shenhav, B., Olender, T., & Lancet, D. (2009). Mutations and lethality in simulated prebiotic networks. Journal of Molecular Evolution, 69(5), 568-578.
- Shenhav, B., Oz, A., & Lancet, D. (2007). Coevolution of compositional protocells and their environment. Philosophical Transactions of the Royal Society B: Biological Sciences, 362(1486), 1813-1819.
- Shenhav, B., Bar-Even, A., Kafri, R., & Lancet, D. (2005). Polymer GARD: computer simulation of covalent bond formation in reproducing molecular assemblies. Origins of Life and Evolution of Biospheres, 35(2), 111-133.
- Segré, D., Shenhav, B., Kafri, R., & Lancet, D. (2001). The molecular roots of compositional inheritance. Journal of Theoretical Biology, 213(3), 481-491.
- Segré, D., Ben-Eli, D., Deamer, D. W., & Lancet, D. (2001). The lipid world. Origins of Life and Evolution of the Biosphere, 31(1), 119-145.
- Segré, D., & Lancet, D. (2000). Composing life. EMBO reports, 1(3), 217-222.
- Segré, D., Ben-Eli, D., & Lancet, D. (2000). Compositional genomes: prebiotic information transfer in mutually catalytic noncovalent assemblies. Proceedings of the National Academy of Sciences, 97(8), 4112-4117.
- Segré, D., Lancet, D., Kedem, O., & Pilpel, Y. (1998). Graded autocatalysis replication domain (GARD): kinetic analysis of self-replication in mutually catalytic sets. Origins of Life and Evolution of the Biosphere, 28(4), 501-514.

