Our Lab research focuses on redox signaling in the plant photosynthetic organelle, the chloroplast. In particular, we are looking at post-transcriptional regulation in the adaptation of photosynthesis to changing environments.
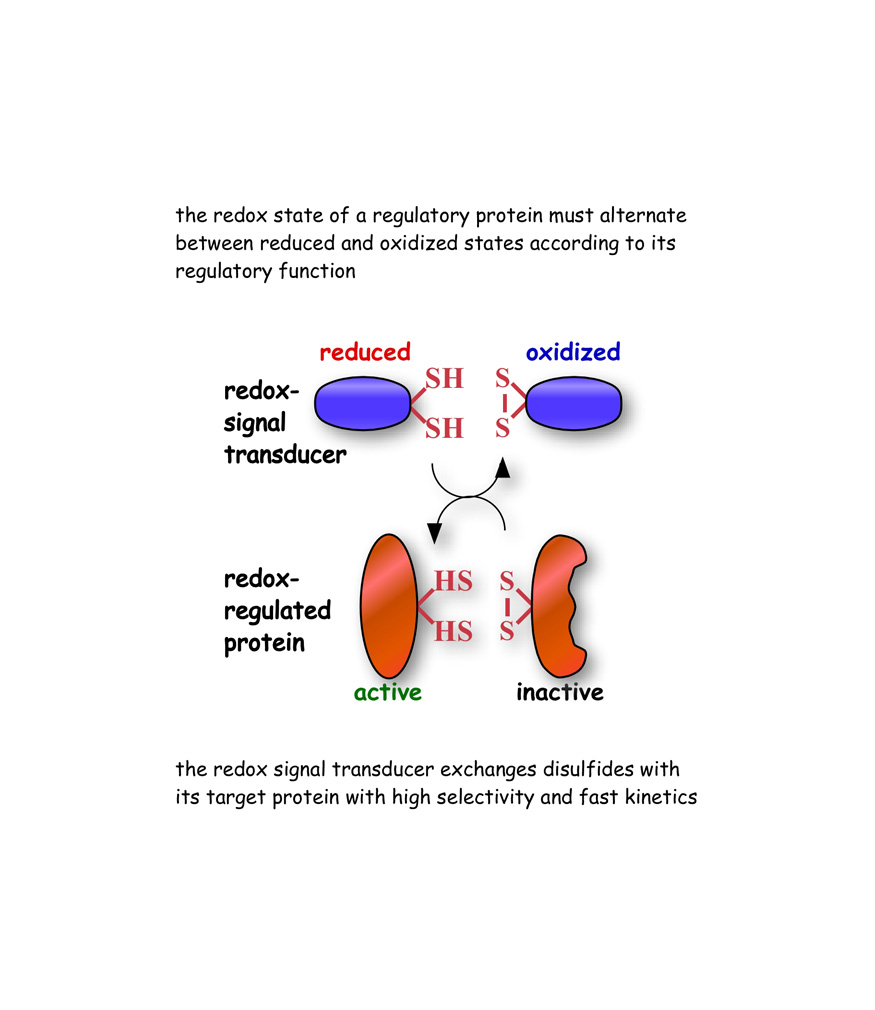
Signaling by redox-active proteins is ubiquitous in all living organisms. However, the molecular mechanisms by which specific redox-active proteins regulate the expression and activity of specific proteins are not well understood. One key question is how redox signals are transmitted and perceived in a specific manner in an intracellular milieu that is highly reductive and buffered against redox changes.
The conversion of light energy into beneficial chemical energy by the photosynthetic machinery also produces under adverse conditions destructive reactive oxygen species. Thus, a tight adjustment of the photosynthetic activity to the changing environmental conditions is required. Interestingly, multiple regulatory mechanisms that directly sense changes in photosynthetic redox signals have evolved. Both reductive type signals, and oxidative type signals are involved in the regulation. These mechanisms allow for a dynamic control that continuously adjusts to changing environmental conditions.
Our research demonstrates that regulatory proteins of the thioredoxin family exchange electrons along specific pathways in the soluble compartments of the cell. Our studies suggest that the flow of electronic information in biology can take place catalytically by ways of high selectivity and fast kinetics properties of enzymatic reactions.
In chloroplasts, multiple redox signaling programs, such as in controlling enzymes of carbon fixation, or in regulation of sunlight captuaring and conversion to chemical energy, or in regulation of protein synthesis, or in protection mechanisms against the accumulation of free radicals, seem to take place in parallel. This raises questions about the identity of the signaling proteins and the principals of their redox reactions.


