Research
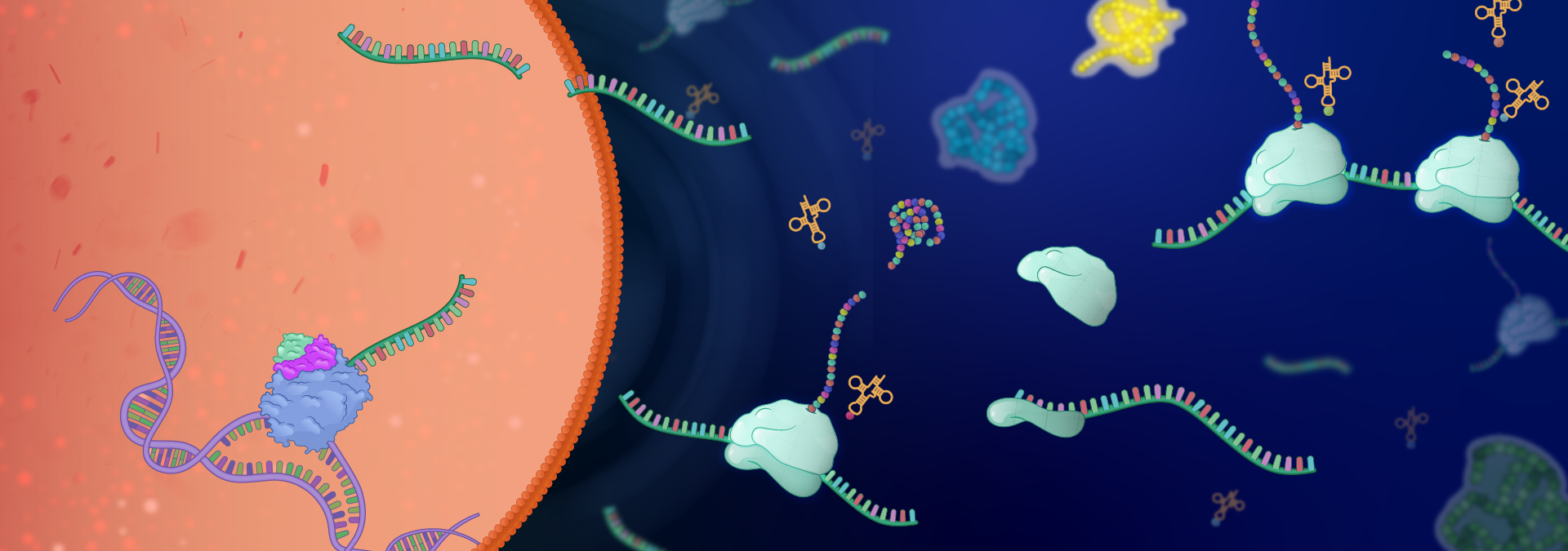
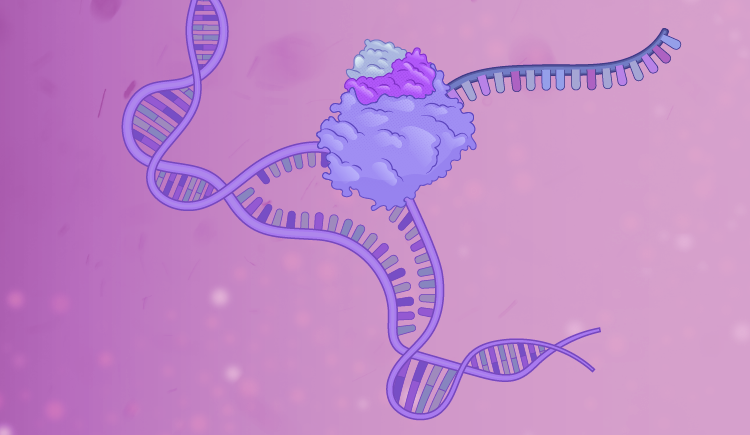
Regulation of gene expression at the transcriptional and translational levels is fundamental to all biological activities and is frequently altered in disease states. Our broad research interest is to elucidate the basic regulatory mechanisms of transcription and mRNA translation. We focus on several key issues:
- How do transcription and translation processes control the response to extracellular stimuli?
- The connections between transcription, mRNA processing and translation.
- The mechanism underlying unique mechanisms of translation initiation.
- Developing chemical and genetic research tools for transcription and translation.
By integrating molecular, biochemical, bioinformatics, and chemical approaches, our studies have led to novel insights related to transcription initiation and elongation, mRNA processing and stability, and translation initiation, as well as the connections between them. We address these issues utilizing several biological systems:
- Spt4/Spt5 (DSIF), a transcription elongation factor involved in stress responses and a prime target for inherited neurodegenerative diseases
- Regulation of translation initiation: (i) the role translation initiation factors (eIFs); (ii) specialized translation driven by ribosome intrinsic mRNA binding (iii) cellular and viral ribosome binding proteins (iv) Stress and mitogenic signals
- Identification of mammalian and viral translation regulatory elements and their potential use in RNA therapeutics
- Mediators of the crosstalk between gene expression stages
Research page