An ensemble of regulatory elements controls Runx3 spatiotemporal expression in subsets of dorsal root ganglia proprioceptive neurons
The phenotypes of Runx3-/- mice reflects Runx3 expression pattern and its vitality to the proper function of several important organs. Among these, Runx3 is essential for development and diversification of the dorsal root ganglia (DRG) TrkC sensory neurons. In Runx3-/- mice, developing TrkC neurons fail to extend central and peripheral afferents leading to cell-death and disruption of the stretch reflex circuit, resulting in severe limb ataxia. Despite its central role, the mechanisms underlying the spatiotemporal expression specificities of Runx3 in TrkC neurons were largely unknown.
Using BACs engineered to express a reporter gene (LacZ) as transgenes, we defined the genomic transcription unit encompassing regulatory elements (REs) that mediate the tissue-specific expression of Runx3 (Figure 1).
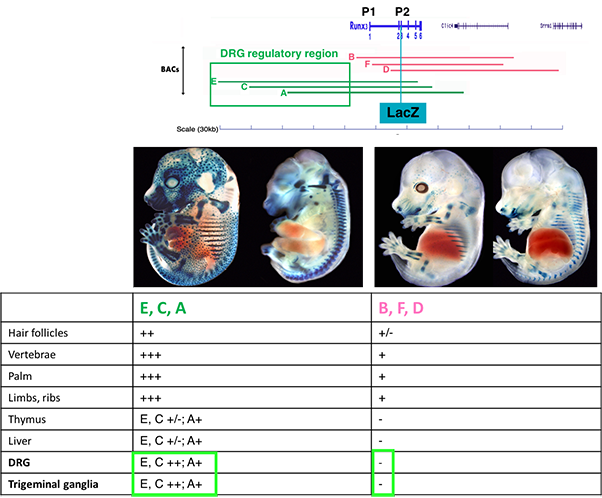
Figure 1 Transgenic mice analysis revealed that BACs extending 5’ upstream of Runx3 (A,C, E,) contain regulatory elements conferring expression in TrkC neurons.
Analysis of conserved non-coding elements included in these BACs led to identification of three REs, dubbed R1, R2 and R3, that crosstalk with promoter-2 (P2) to drive TrkC neuronspecific Runx3 transcription. Deletion of single or multiple elements either in the BAC transgenics, or by CRISPR/Cas9-mediated endogenous ablation, established the REs' ability to promote and/or repress Runx3 expression in developing sensory neurons (Figure 2). Our analysis reveals that an intricate combinatorial interplay among the three REs drives Runx3 expression in distinct subtypes of TrkC neurons, while concomitantly extinguishing its expression in non-TrkC neurons. These findings provide new insights into the mechanism regulating cell type-specific expression and subtypes diversification of TrkC neurons in developing DRG.
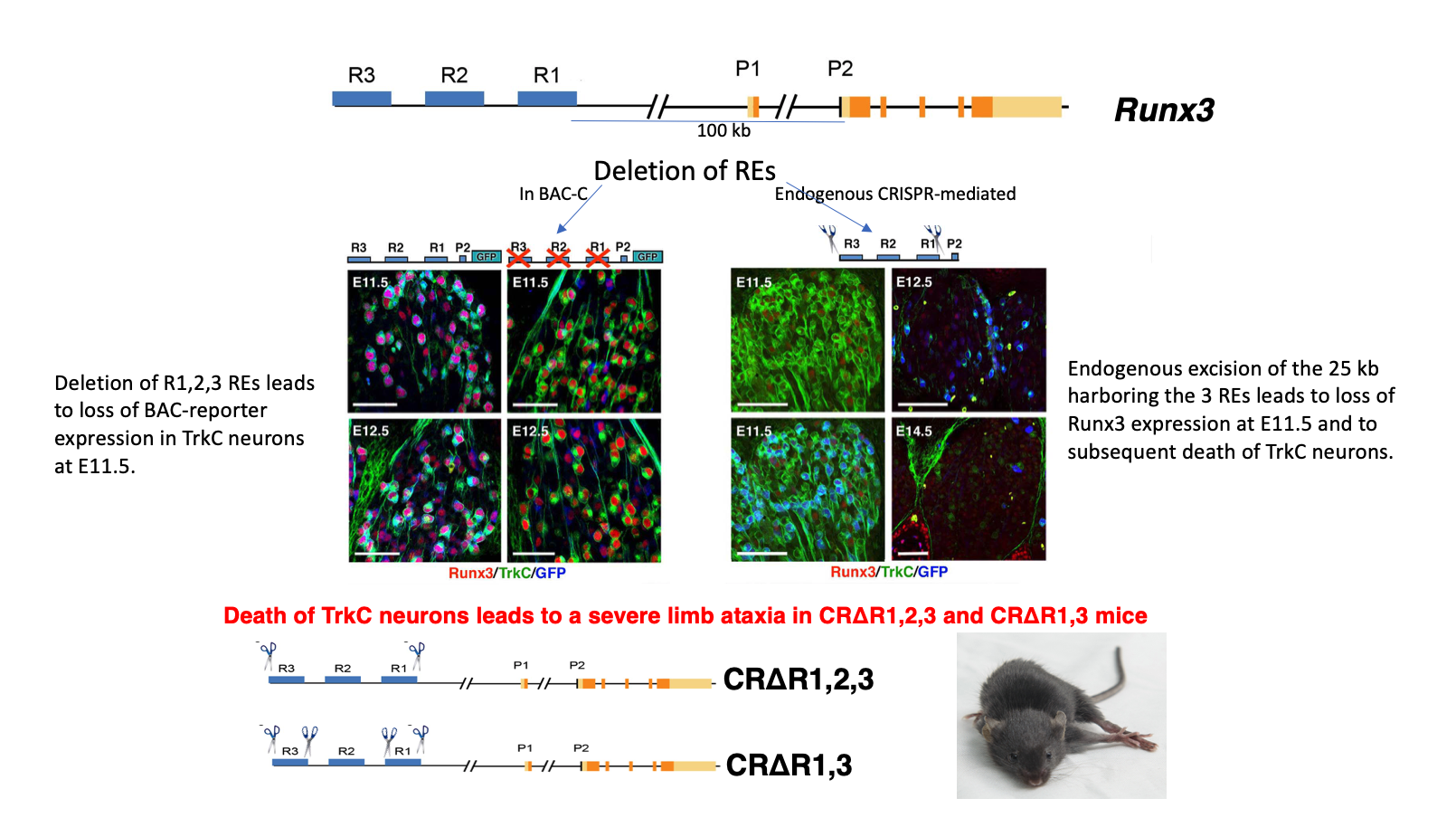
Figure 2 The R1, R2 and R3 regulatory elements located 100-125Kb upstream of the P2 promoter confer Runx3 expression in TrkC neurons (BAC deletions and endogenous CRISPR mediated deletions of REs)
Identification of Runx3 targets in TrkC neurons
To identify of Runx3 target genes in developing TrkC neurons, we conducted RNA-seq using RNA from FACS isolated TrkC/GFP neurons of WT/P2GFP and Runx3-/- i.e. homozygous P2GFP/GFP E11.5 embryos. A total of 466 genes were Runx3 responsive, exhibiting differential expression in the absence of Runx3 (P2GFP/GFP / WT/P2GFP ≥ 2; FDR< 0.05). Of these, 338 genes were upregulated and 128 genes were downregulated in the absence of Runx3. Among these genes those depicted in Figure 3 changed in the same direction both in Pou4f1-/- and Runx3-/- TrkC neurons, indicating that these two TFs cooperate in regulating of expression of these genes.
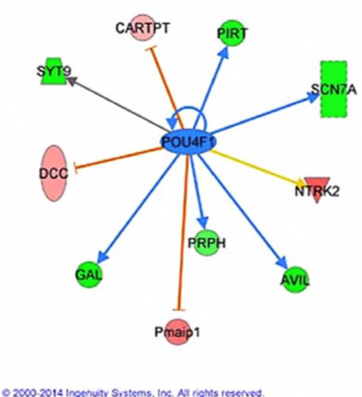
Figure 3 Several genes identified as Pou4f1 targets are also Runx3 target genes (green marks downregulated genes and pink marks upregulated genes).
ChIP-seq with anti Runx3 could sort out the differentially expressed genes that are direct target genes of Runx3. However, currently most protocols for TF ChIP-seq require isolation of large number of TrkC neurons, a task infeasible today. To gain more information on Runx3 transcription regulation, we established the genomic profiles of chromatin modifiers, H3K27ac and H3K4me1as well as ATAC peaks on chromatin of E11.5 DRG TrkC neurons.