DAP5 and Cap-Independent Protein Translation
DAP5 (EIF4G2) is a translation initiation factor of the EIF4G family, which directs non-canonical, mostly cap-independent, protein translation. It provides a mechanism to maintain translation of itself and specific target mRNAs under stress circumstances when overall cap-dependent translation is compromised, thereby contributing to the regulation of a cell’s life-or-death decisions, such as during mitosis and ER stress. DAP5 mediates translation initiation by several mechanisms, including recruitment of the translation initiation complex to internal ribosome entry sites (IRES), undefined structured regions within the 5’UTR. A second mechanism involves targets with long 5’UTRs containing upstream open reading frames (uORFs). These small uORFs are translated in a cap-dependent manner independently of DAP5, and their presence generally reduces translation of the main coding sequence (CDS). However, by facilitating read-through of the uORFs and/or re-initializing translation following their translation termination, DAP5 promotes initiation at the CDS, thereby enabling efficient translation of these targets.
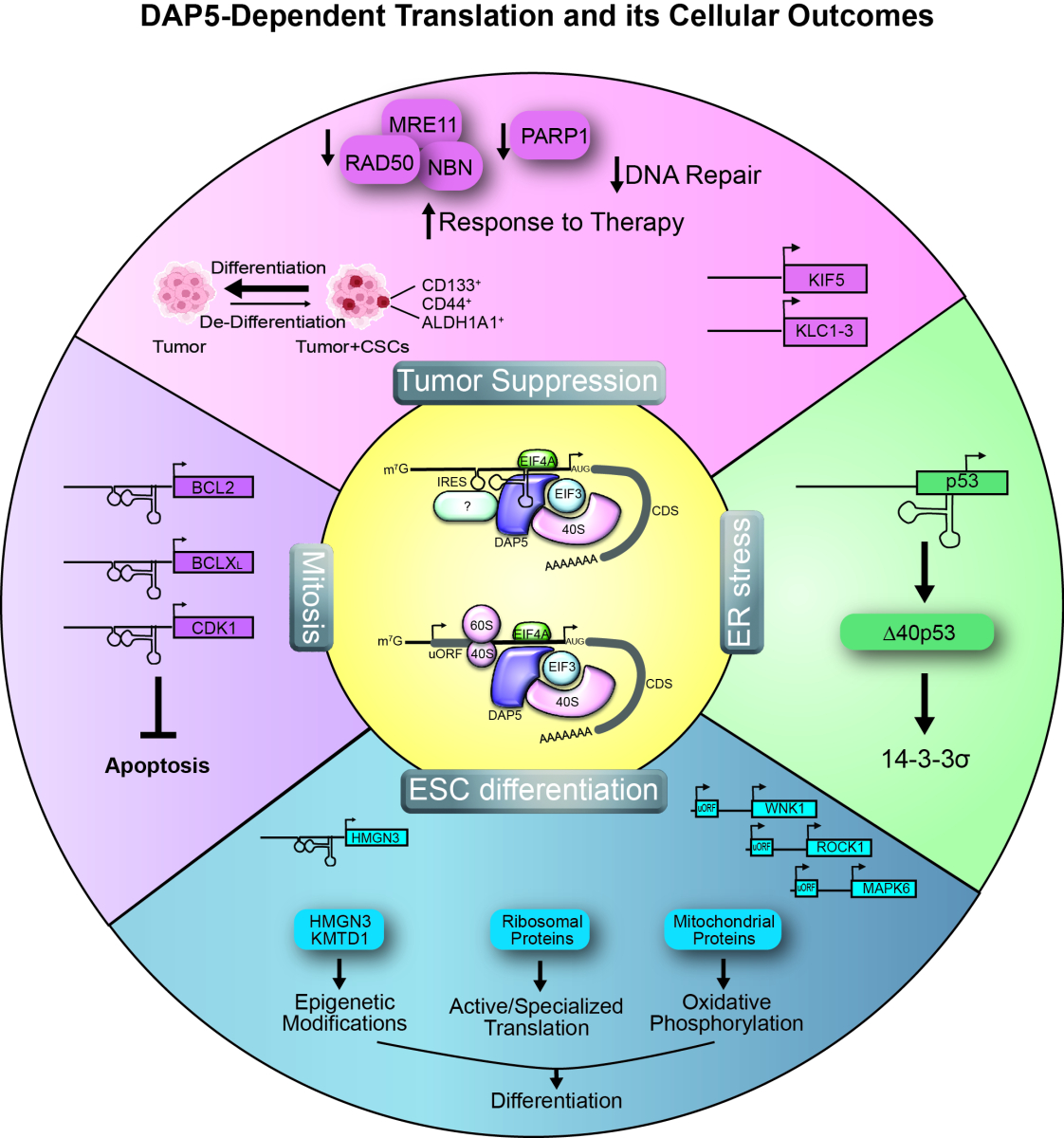
DAP5 can drive translation initiation through IRES elements located upstream to the start codon, or by facilitating translation of the CDS downstream of uORFs (center). Using these mechanisms to drive translation of the specific targets depicted, it determines different cellular outcomes. For more details, please see our papers: Tumor suppression: Meril, et al. 2024 Oncogene 43:1098-111; ESC differentiation: Yoffe, et al, 2016, Genes Dev. 30:1991-2004, David, et al., 2022, RNA 28:1325-1336; ER stress: Weingarten-Gabbay, et al. 2014 Oncogene. 33: 611-618; and Mitosis: Marash, et al. 2008 Mol Cell. 30:447-459, Liberman, et al. 2009 Cell Cycle. 8:204-209.
Translation control of embryonic stem cell differentiation
DAP5 is necessary for human embryonic stem cell (hESC) differentiation, and a major direction in our lab in recent years is to understand how DAP5’s function as a non-canonical translation factor regulates this process. We used a multi-pronged approach to identify DAP5 target mRNAs, including RNA-seq of mRNAs associated with heavy polysomal fractions (polysome profiling) or occupied by ribosomes (ribosome profiling). We also examined the DAP5-mRNA interactome by identifying mRNAs that co-IP with DAP5 upon cross-linking in hESCs (CLIP). The results of these strategies are summarized in the above scheme.
In addition to DAP5, we are currently investigating PRRC2B, an RNA binding protein that strongly interacts with DAP5, with the aim of identifying its roles in differentiation and development. PRRC2B is a large, disordered protein that has been previously implicated as an M6A reader, and is also involved in translation initiation, specifically of mRNAs with uORFs. We have discovered that PRRC2B is necessary for hESC differentiation to specific cell lineages. Currently, a major focus in the lab is to investigate PRRC2B’s function in translation, and to identify its complete interactome and target mRNAs. In parallel, we are analyzing the consequence of PRRC2B knock-out on mouse development, to establish the physiologic outcomes of its differentiation functions.
Translation control of cancer development
Recently, we have expanded our studies to cancer stem cells, to determine whether DAP5 may contribute to malignancy by regulating the pluripotency/differentiation balance in somatic stem cells as well, thereby acting as a tumor suppressor. Cancer stem cells (CSCs) are present in small numbers in various solid tumors, representing a pool of cells that is more malignant and drug resistant than the more differentiated general cancer cell population. We have shown that DAP5 depletion in endometrial cancer (EC) cells is associated with a more aggressive, stem cell-like phenotype, and leads to a significantly altered transcriptional and proteomic landscape. A significant increase in abundance was observed for several DNA damage repair proteins, and our results indicated that KD cells are better equipped to resolve DNA damage and are more likely to survive DNA damaging treatments. Prominent among the proteins with decreased abundance were Kinesin-1 motor proteins, including the heavy chain KIF5B and light chains KLC1, 2, 3. In addition, we conducted Kaplan-Meier survival analysis and multiplex tumor microarray immunostaining for DAP5 protein and its putative targets in a cohort of 280 EC patients. Significantly, we found that low expression of DAP5 and kinesin proteins highly correlated with poor overall- and recurrence-free survival in patients with tumors of certain grades and stages.
In parallel, we have mined the COSMIC tumor database for DAP5 non-syngeneic somatic mutations, to further explore the contribution of DAP5 to cancer. Structure-function studies of these mutants identified specific DAP5 variants with impaired ability to drive translation initiation and interact with DAP5 binding partners. The presence of these loss-of-function mutations in cancers, and their prevalence in endometrial cancer specifically, is strong support for a tumor-suppressive role for DAP5.
A. B.
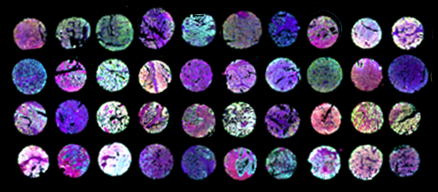
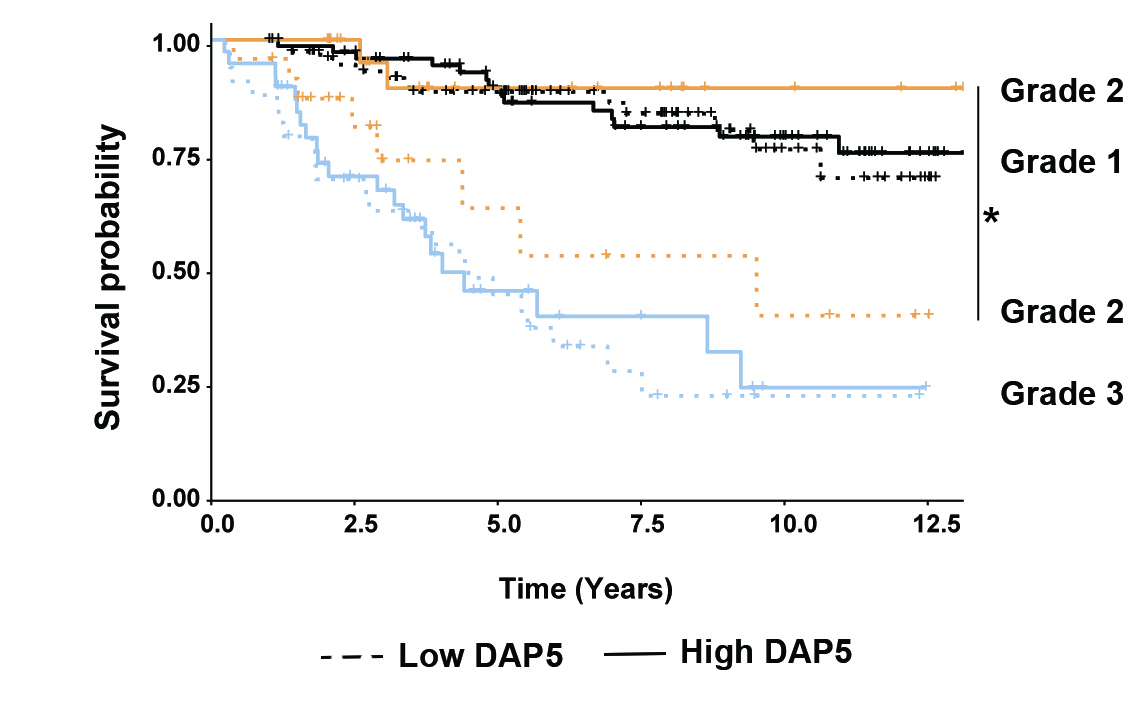
A. Multiplex staining of tumor array containing cores from EC patients. B. Kaplan-Meier Survival analysis of EC patients, stratified according to tumor grade and DAP5 expression. Adapted from Meril, et al., 2024 Oncogene 43:1098-111.


